S1 = MEGKEENMR S2 = MVGKERR S3 = NVGRIINMV S4 = NVRINMV
Let the scoring function s(x,y) be
| s(x,y)=0 | if x matches (equals) y; |
| s(x,y)=1 | if x does not match (equal) y; |
| s(x,y)=1 | if either x or y is space (-), but not both. |
Use the center star method to find an approximately optimal SP alignment for the four strings given. Show all your computations.
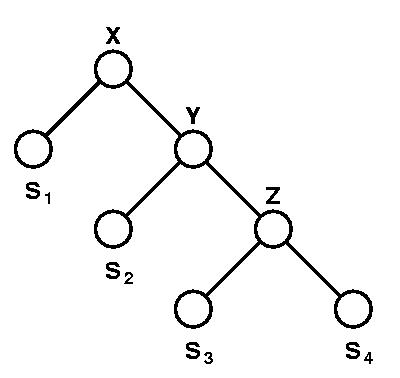
Using the approximation approach in Gusfield 14.8.1, compute an optimal lifted alignment for T. What is the distance of your alignment?
Additional Challenge for the Bored
(Not required; no extra credit offered)
Find a phylogenetic tree whose optimal phylogenetic alignment
(Gusfield 4.8) for S1, S2,
S3, and S4
has minimum distance.
In particular,
the best phylogenetic tree may have no structural similarity to T.